Colony Formation Assay Quantification

Figure 1 From Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays Semantic Scholar
Clonogenic Assay Creative Bioarray Creative Bioarray
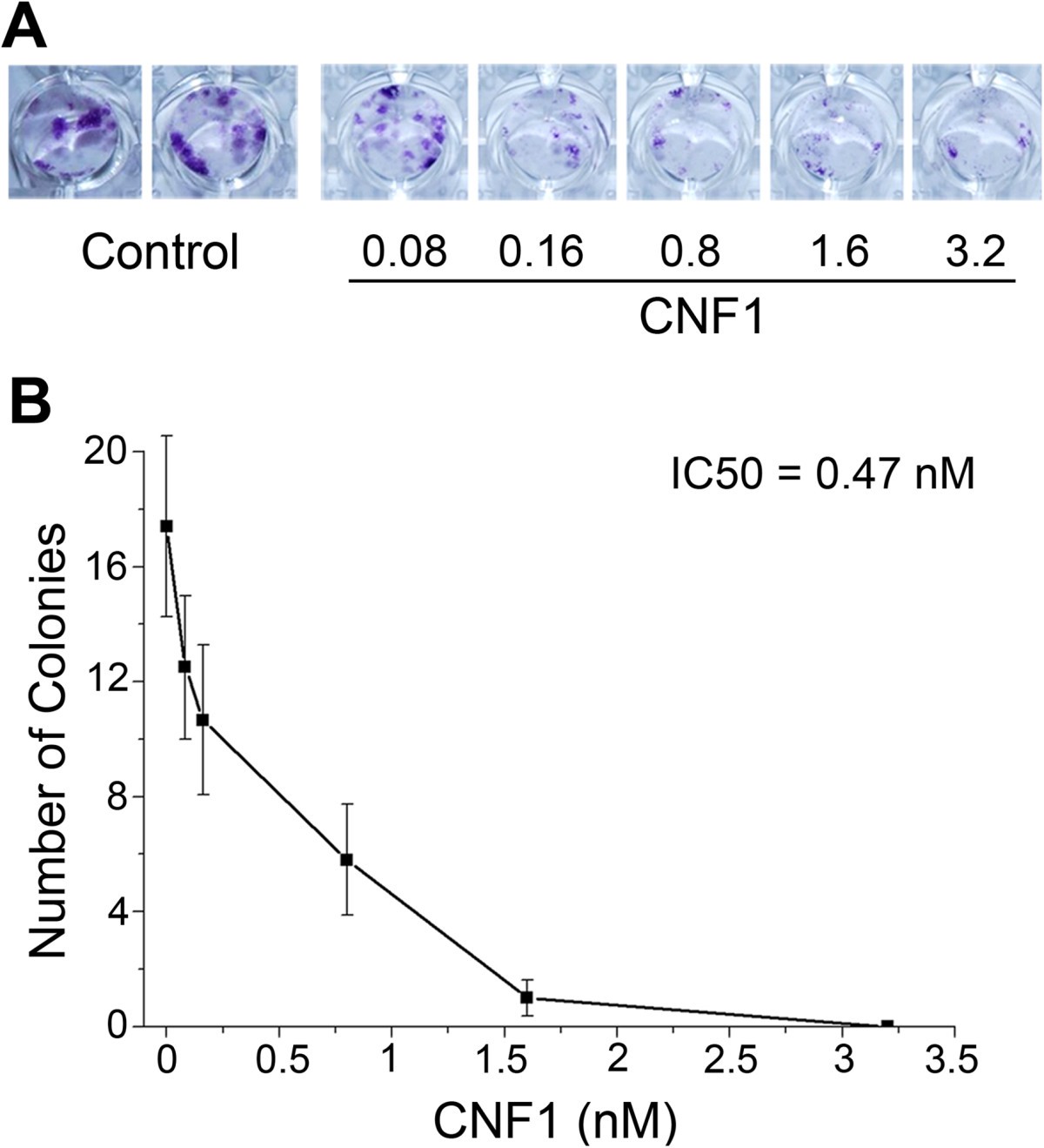
The Bacterial Protein Toxin Cytotoxic Necrotizing Factor 1 Cnf1 Provides Long Term Survival In A Murine Glioma Model Bmc Cancer Full Text

Mir 2b Silencing Activates Nf Kb And Promotes Aggressiveness In Breast Cancer Cancer Research

Cmpd1 Inhibited Human Gastric Cancer Cell Proliferation By Inducing Apoptosis And G2 M Cell Cycle Arrest

Figure 2 From Chloroform Fraction Of Serratulae Chinensis S Moore Suppresses Proliferation And Induces Apoptosis Via The Phosphatidylinositide 3 Kinase Akt Pathway In Human Gastric Cancer Cells Semantic Scholar
Thus, the researcher must measure the diameter of each colony (21).
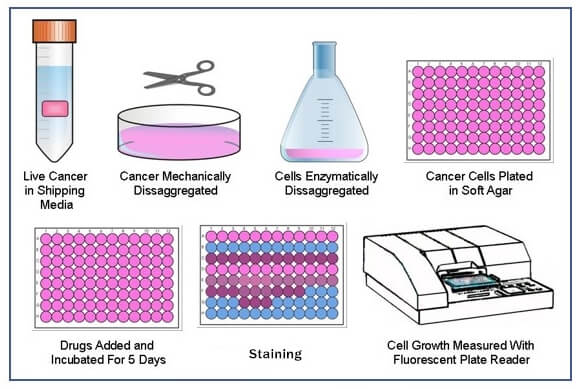
Colony formation assay quantification. One to two weeks are sufficient for HeLa cells to form colonies that are easy to count The time required to grow colonies from other cell types will vary based on the proliferation rates of each individual cell type Colonies of ∼ 50 cells are sufficient for counting by light microscopy at low resolution. Lay et al 05) or genome probing with micro arrays (Bae et al 05) have gained popularity (Liu et al 04a). The primary method of monitoring anchorageindependent growth is the detection of soft agar colony formation, which measures proliferation by manual counting of colonies in semisolid culture media This traditional assay, though effective, is rather laborious in the initial setup and final quantification stages.
The Soft Agar Assay for Colony Formation is an anchorage independent growth assay in soft agar, which is considered the most stringent assay for detecting malignant transformation of cells For this assay, cells (pretreated with carcinogens or carcinogen inhibitors) are cultured with appropriate controls in soft agar medium for 2128 days. Subsequent counting of colony forming units (CFUs) In recent years a range of alternative, highthroughput (HT), methods relying on quantitative PCR (Neeley et al 05;. Most measurements of attachment have used microscopy and colony counting, both of which are labor and time intensive To reduce the time and effort required to analyze bacteria attaching to plant tissues, we developed a quantitative realtime PCR (qPCR) assay to quantify attached A tumefaciens using the chvE gene as marker for the presence of the bacteria.
Lay et al 05) or genome probing with micro arrays (Bae et al 05) have gained popularity (Liu et al 04a). The colony forming cell (CFC) assay, also referred to as the methylcellulose assay, is an in vitro assay used in the study of hematopoietic stem cells The assay is based on the ability of hematopoietic progenitors to proliferate and differentiate into colonies in a semisolid media in response to cytokine stimulation. Thus, the researcher must measure the diameter of each colony These techniques are necessarily subjective since the researcher has to manually trace the regions of interest in.
Crystal Violet Cell Colony Staining 1L Fixing/Staining solution 05 g Crystal Violet (005% w/v) 27 ml 37% Formaldehyde (1%) 100 mL 10X PBS (1X) 10 mL Methanol (1%) 863 dH to 1L 1) Remove media (do not wash cells) 2) Add staining solution to cover dish 3) Stain for min at room temperature 4) Remove fix/stain solution and save. A Representative images of the colony formation assays using MCF7 or MCF7/TAMR cells under 0, 5, and 10 μM tamoxifen treatment for 14 days The bar graphs show the quantification of the colony formation assay data The data are presented as the mean ± SD of three independent experiments Student’s t test was used for statistical analysis. Vitro or in vivo assays 8;.
In summary, our colony assay allows easy detection and quantification of functional progenitors within a heterogeneous population of cells In addition, the semisolid media format allows uniform presentation of extracellular matrix components and growth factors to cells, enabling progenitors to proliferate and differentiate in vitro. Recently advances have been made in the soft agar colony formation assay to increase the speed and accuracy of quantitation These assays utilize a semisolid agar medium in which colonies are quantified using a fluorometric dye, thereby eliminating manual counting. A clonogenic assay, also known as a colony formation assay is an in vitro cell survival assay It assesses the ability of single cells to survive and reproduce to form colonies 1 This assay was first described in the 1950s, where it was used to study the effects of radiation on cancer cell survival and growth and has subsequently played an essential role in radiobiology 2.
Cell colony as indicated A 50cell colony for Caco2 and HeLa cells corresponds to an area of 103x106 and 90x105 mm2, respectively These values were applied to color brightfield images of Crystal Violetstained colonies (C and F) Scale bar = 0 μm Automated Color Brightfield Imaging and Analysis of the Colony Formation Assay. It is necessary to convert the quantification cycle (Cq) value from the qPCR assay to colony forming units (CFU) State regulations for microbial thresholds currently use nomenclature from culturebased methods to set acceptable thresholds With this in mind, it was necessary for Medicinal Genomics to convert the quantification cycle (Cq) value from the PathoSEEK® qPCR Assay to colony forming units (CFU), which is reported when using culturebased methods. Haarman and Knol 06), fluorescent labelling (Blasco et al 03;.
Clonogenic assay or colony formation assay is an in vitro cell survival assay based on the ability of a single cell to grow into a colony The colony formation assay is a sensitive measure of compounds which requires cells to undergo several rounds of replication in order for the effect to be observed. 321 Colonyforming unit assays CFU assays were first described in 1966 by Bradley and Metcalf as the shortterm quantitative assay for the measurement of differentiative potential of lineagerestricted progenitor cells (Bradley and Metcalf, 1966). The ‘gold standard’ for quantification of cytotoxicity is the colony forming assay This assay requires that colonies grow long enough to form visible colonies that are then counted manually We have created a new approach that enables miniaturization and automation.
Soft Agar Colony Formation Assay Colony formation in soft or hard agar Last updated 8/12/14 By Madison Weg and Tim Starr Overview Use this protocol to test for cellular transformation exhibited by the ability to grow in an anchorageindependent setting Normal cells will not grow in soft agar due to anoikis, while transformed cells will. Endothelial progenitor cells (EPCs) play a critical role in restoration of ischemic diseases However, the actual status of EPC development and the mechanisms of EPC dysfunctions in patients with various ischemic diseases remain unknown To investigate the detailed function of EPCs in experimental murine models, we have established an EPC colony forming assay (EPCCFA) in murine EPCs. Measurement of colonies made of fluorescently labeled cells is also an option;.
The bicinchoninic acid assay (BCA) is based on a simple colorimetric measurement and is the most common protein quantification assay BCA is similar to the Lowry or Bradford protein assays and was first made commercially available by Pierce, which is now owned by Thermo Fisher Scientific. In particular, the colony formation assay has become a standard experiment for characterizing the tumor development in vitro However, quantification of the growth of cell colonies under a microscope is difficult because they are suspended in a threedimensional environment. This can be achieved using the colonyforming assay described here This protocol specifically applies to measurement of HeLa cells but can be used for most adherent cell lines with limited motility.
Clonogenic assay or colony formation assay is extensively used to measure in vitro cell survival based on the capacity of a single cell to grow into a colony When a cell divides and form a cluster of more than 50 cells, known as colony, is regarded as viable and traditionally counted manually 1 To obtain statistical accuracy such assays. The primary method of monitoring anchorageindependent growth is the detection of soft agar colony formation, which measures proliferation by manual counting of colonies in semisolid culture media This traditional assay, though effective, is rather laborious in the initial setup and final quantification stages. One to two weeks are sufficient for HeLa cells to form colonies that are easy to count The time required to grow colonies from other cell types will vary based on the proliferation rates of each individual cell type Colonies of ∼ 50 cells are sufficient for counting by light microscopy at low resolution.
(C) Scans of representative MCF10A colony formation assays (D) Colony formation assay quantification of PMAtreated (25 ng/mL, 6 h) MCF10A cells relative to control (DMSO) Bars represent the average of 3 biological replicates ± SEM. Protein quantification using Bradford Assay with Coomassie Overview Protein binds to the Coomassie Plus reagent and causes a shift in signal from 465 to 595 nm with a simultaneous color change of the reagent from green/brown (or red/brown) to blue Pierce Coomassie Plus Protein Assay (Cat # ) has a detection range of 1µg/ml to 1µg/ml. Seventeen different L monocytogenes strains were tested using a modified crystal violet assay for their quantification The evaluation of their biofilmforming ability was carried out after a 48hour incubation period at 30ºC The media employed was the one proposed by Pan et al, 31 which was TSYEB with a supplement of glucose and sodium chloride.
004% DMSO) Average colony numbers were quantified and are graphed as mean (±SD) from triplicate wells (*** p Techniques Used Inhibition, In Vitro, In Vivo. The ‘gold standard’ for quantification of cytotoxicity is the colony forming assay This assay requires that colonies grow long enough to form visible colonies that are then counted manually We have created a new approach that enables miniaturization and automation. Quantification of colony formation measuring absorption Colony formation was quantified following the method described by Kueng and coworkers In brief, the crystal violet staining of cells from each well was solubilized using 1 ml of 10% acetic acid and the absorbance (optical density) of the solution was measured on a Synergy H1 hybrid fluorescence platereader (BioTek, Winooski, VT, USA) at a wavelength of 590 nm.
Subsequent counting of colony forming units (CFUs) In recent years a range of alternative, highthroughput (HT), methods relying on quantitative PCR (Neeley et al 05;. These data can also be used to calculate mean cells per colony Traditional methods for quantification of colonies by handcounting coupled with an assay for cell number (for example, DNA or mitochondrial) remains a viable method that can be used to calculate the mean number of cells per colony. Endothelial progenitor cells (EPCs) play a critical role in restoration of ischemic diseases However, the actual status of EPC development and the mechanisms of EPC dysfunctions in patients with various ischemic diseases remain unknown To investigate the detailed function of EPCs in experimental murine models, we have established an EPC colony forming assay (EPCCFA) in murine EPCs.
Protein quantification using Bradford Assay with Coomassie Overview Protein binds to the Coomassie Plus reagent and causes a shift in signal from 465 to 595 nm with a simultaneous color change of the reagent from green/brown (or red/brown) to blue Pierce Coomassie Plus Protein Assay (Cat # ) has a detection range of 1µg/ml to 1µg/ml. The primary method of monitoring anchorageindependent growth is the detection of soft agar colony formation, which measures proliferation by manual counting of colonies in semisolid culture media This traditional assay, though effective, is rather laborious in the initial setup and final quantification stages. Analyze the whole well continuously.
In summary, our colony assay allows easy detection and quantification of functional progenitors within a heterogeneous population of cells In addition, the semisolid media format allows uniform presentation of extracellular matrix components and growth factors to cells, enabling progenitors to proliferate and differentiate in vitro. The Soft Agar Assay for Colony Formation is an anchorage independent growth assay in soft agar, which is considered the most stringent assay for detecting malignant transformation of cells For this assay, cells (pretreated with carcinogens or carcinogen inhibitors) are cultured with appropriate controls in soft agar medium for 2128 days. Quantify the ability of a single cell to form a colony A labelfree assayuse bright field imaging to identify the formation of a singlecell clone colony;.
In many assays biofilms are quantified by conventional culture plating method to get colony 10 forming units/count, which is an intensive procedure 1 Whereas other assays do use 96 well 11 microtiter plates for biofilm quantification as microtitre plate offers comparatively high 12. Haarman and Knol 06), fluorescent labelling (Blasco et al 03;. For the softagar assay, the formula commonly employed is A = πRr, where π = 314, and R and r are the longest and the shortest radii, respectively, of the colonies;.
‘‘ColonyArea’’, which is optimized for rapid and quantitative analysis of focus formation assays conducted in 6 to 24well dishes ColonyArea processes image data of multiwell dishes, by separating, concentrically cropping and background correcting well images individually, before colony formation is quantitated. The colonyforming unit (CFU) assay is one of the most widely used assays for hematopoietic stem and progenitor cells (HSPCs) CFU assays allow measurement of the proliferation and differentiation ability of individual cells within a sample The potential of these cells is measured by the observation of the colonies (consisting of more differentiated cells) produced by each input progenitor cell. True cellpercolony quantification, no areapercolony estimation needed;.
9 In vitro Tube Formation Assay This assay involves plating endothelial cells onto a basementmembranelike substrate on which the cells form tubules within six to hours These tubules mostly contain a lumen, and the cells develop tight cellcell and cellmatrix contacts Quantification can be. One particular assay where livecell imaging can be useful is the colony formation assay This assay determines a cell’s ability to form a colony over a long period of time in culture 6 However, colony counting and assessment of colony sizes are primarily done manually, which can be a tedious process 6 To support researchers specific software developed for livecell imaging enables quantification of colony formation (colony size and colony count) over time, reducing manual labor. A colony assay that allows functional and quantitative assessments of single progenitor cells Numeration of the resulting colonies is used to calculate the frequency of the progenitor cells among the total plated cells The composition of cells within each colony is indicative of the lineage potential of the initiating progenitor.
The initial cell number of each microorganism was determined by colony counting Each microorganism cell culture was diluted with medium and 190 μl of the cell culture was added to each well Then 10 μl of assay solution was added. Regulations for microbial threshold tests like TYM currently use nomenclature from culturebased methods to set acceptable thresholds It is necessary to convert the quantification cycle (Cq) value from the qPCR assay to colony forming units (CFU). Indeed, addition of exogenous VEGF to OP9 culture induces dispersion of the EC colony, rendering the quantification of the EC colony difficult 12 Nonetheless, the OP9 cell line provides a suitable condition for enumerating the number of clonogenic EC progenitors from both ES cell differentiation cultures and the actual embryos Because the proliferative capacity of EC may decrease on maturation, it should be noted that the proportion of EC progenitors clonable by this assay would decrease.
Final colonyforming units (CFUs) are calculated by correcting for the dilution factor 33 Viability Measured by FUN1 Staining Before heat ramp treatment ( see Note 7 ), 05 μl FUN1 cell stain and 5 μl Calcofluor White M2R are added to 1 ml yeast culture (OD 600 adjusted) with final concentrations of 5 and 25 μM, respectively. Next, we investigated the effect of SUV39H1 silencing on EVderived miR744 mediated NSCLC cell proliferation using CCK8 assay and colony formation assay The results showed that the proliferation and colony formation in BEAS2BEVtreated A549 and H460 cells were retarded by siSUV39H11 treatment. Assay parameters can be easily adjusted to accommodate different metabolic states and strain backgrounds Highthroughput BioSpot counting technology with image recognition software adds additional power to this assay The BioSpot Analyzer counts up to 500 microscopic colonies per spot in a 96place format.
Colony Formation Human bone marrow derived CD34 Hematopoietic Progenitor Cells were seeded at 3000 cells/well and cultured for 7 days in the presence or absence of growth factors/cytokines (hIL3, hIL6, hGCSF, hEPO) Colony quantitation was determined according to the assay protocol. In particular, the colony formation assay has become a standard experiment for characterizing the tumor development in vitro However, quantification of the growth of cell colonies under a microscope is difficult because they are suspended in a threedimensional environment. I’m trying to perform the colony formation assay (or clonogenic assay) with a cancer cell line to study the effect of DNA repair inhibitors on the cytotoxicity of chemotherapeutic drugs.
Anchorageindependent growth of U251 cells was tested using colony formation assays in soft agar, in the presence of K145 (a), MPA08 (b) and ABC (c) or vehicle control (veh;. In cancer research, colony formation assay is a gold standard for the investigation of the development of early tumors and the effects of cytotoxic agents on tumors in vitro Quantification of cancer cell colonies suspended in hydrogel is currently achieved by manual counting under microscope It is challenging to microscopically quantify the colony number and size without subjective bias. MethoCult™ is the “Gold Standard” for the in vitro detection and quantification of hematopoietic progenitor cells in the colonyforming unit (CFU) or cell (CFC) assay, also known as the methylcellulose assay A wide range of MethoCult™ media formulations are available for CFU assays with hematopoietic cells from human and mouse tissues.
For the softagar assay, the formula commonly employed is A = πRr, where π = 314, and R and r are the longest and the shortest radii, respectively, of the colonies;. Recently advances have been made in the soft agar colony formation assay to increase the speed and accuracy of quantitation These assays utilize a semisolid agar medium in which colonies are quantified using a fluorometric dye, thereby eliminating manual counting. The colony formation assay is an essential method for cancer research, enabling drug screens and radiation dosing to be conducted 15 The assay is performed by seeding cells at a low enough density such that individual cells can propagate to a sufficient colony area without impinging on a neighboring colony (Figure 1) 6, 7 At a set time point, adherent colonies are fixed then stained with Crystal Violet colorimetric dye, which allows for visual inspection of the culture vessel and.
A colonyforming unit ( CFU, cfu, Cfu) is a unit used in microbiology to estimate the number of viable bacteria or fungal cells in a sample Viable is defined as the ability to multiply via binary fission under the controlled conditions Counting with colonyforming units requires culturing the microbes and counts only viable cells, in contrast with microscopic examination which counts all cells, living or dead. A cell viability reagent is added, and fluorescence is measured as an indirect quantification of colony growth in soft agar using Resazurin (D) Example Data The Aurora B inhibitor VX680 was tested for inhibition of the soft agar growth of A549 nonsmall cell lung cancer cells at indicated concentrations.

Utilization Of The Soft Agar Colony Formation Assay To Identify Inhibitors Of Tumorigenicity In Breast Cancer Cells Protocol

Clonogenic Assay A B16 F10 B 058 C Jr8 Cells Were Treated Download Scientific Diagram

Tumor Spheroid Formation Assay Sigma Aldrich

Biofilm Formation Assay In Pseudomonas Syringae

Growth Of Epithelial Organoids In A Defined Hydrogel Broguiere 18 Advanced Materials Wiley Online Library

Comparison Of The Colony Formation And Crystal Violet Cell Proliferation Assays To Determine Cellular Radiosensitivity In A Repair Deficient Mcf10a Cell Line Sciencedirect
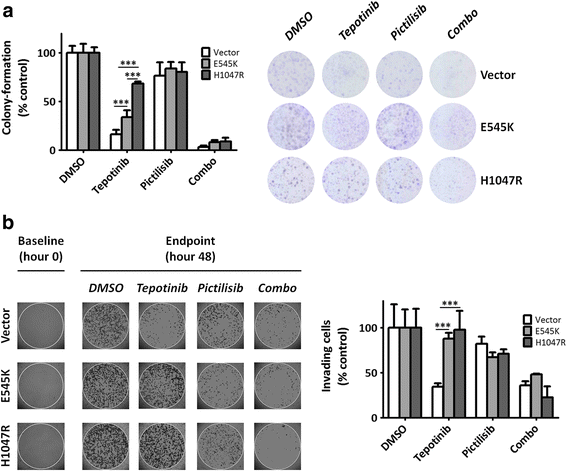
Pik3ca Hotspot Mutations Differentially Impact Responses To Met Targeting In Met Driven And Non Driven Preclinical Cancer Models Molecular Cancer Full Text

An Hts Compatible 3d Colony Formation Assay To Identify Tumor Specific Chemotherapeutics Semantic Scholar

Tuft1 Promotes Osteosarcoma Cell Proliferation And Predicts Poor Prognosis In Osteosarcoma Patients In Open Life Sciences Volume 13 Issue 1 18

High Throughput Fluorescent Colony Formation Assay January 10
Plos One Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays

High Throughput Fluorescent Colony Formation Assay January 10
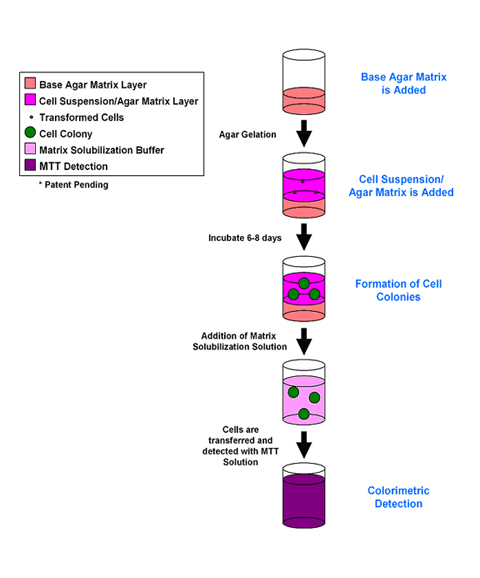
Tumor Sensitivity Assay
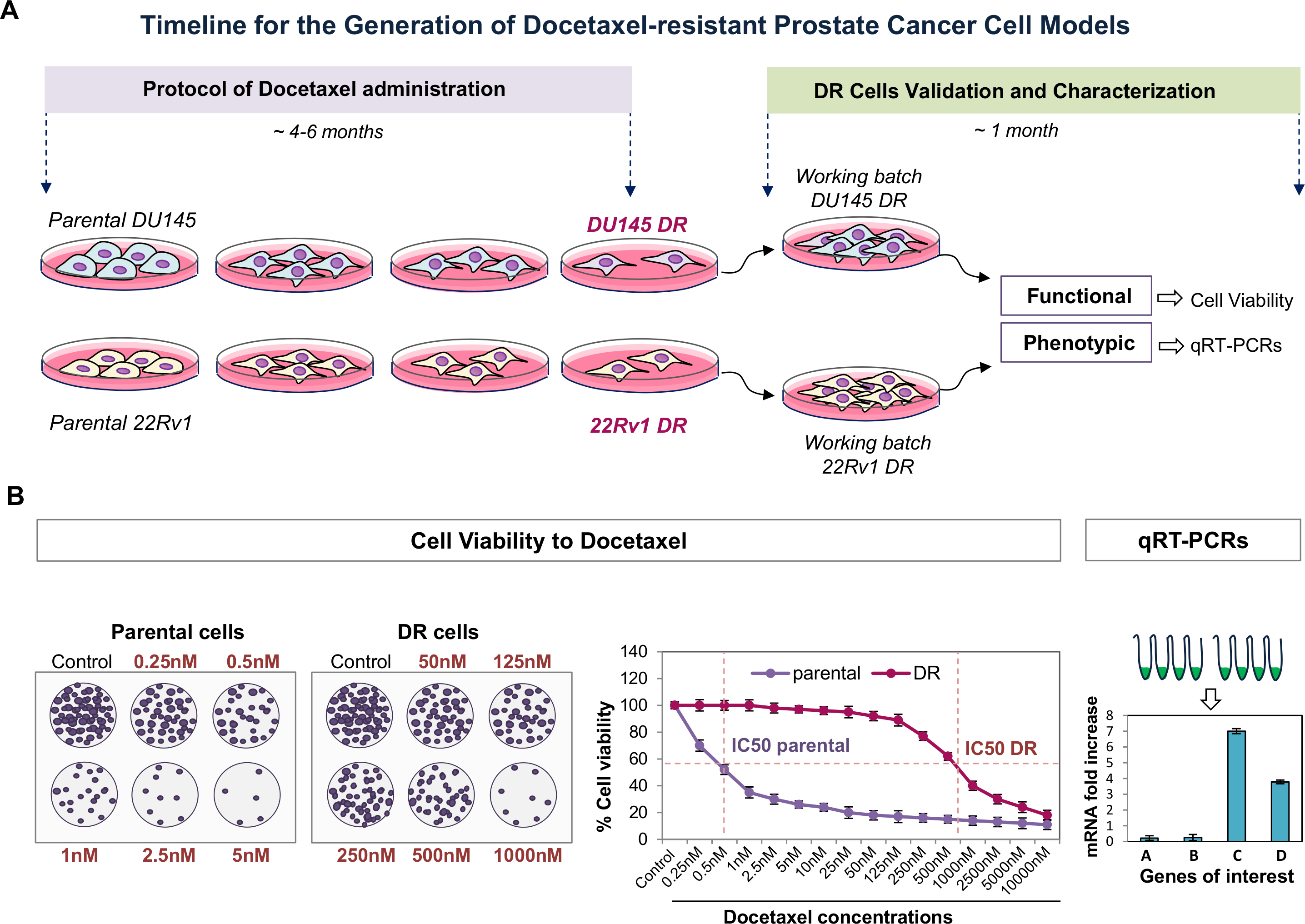
Generation Of Prostate Cancer Cell Models Of Resistance To The Anti Mitotic Agent Docetaxel Protocol

Preclinical Assessment Of The Bioactivity Of The Anticancer Coumarin Ot48 By Spheroids Colony Formation Assays And Zebrafish Xenografts Protocol

Oncogenic Transformation Of Human Mammary Epithelial Cells By Autocrine Human Growth Hormone Cancer Research

High Throughput Fluorescent Colony Formation Assay Metrolab Blog

Colony Forming Assay Verification Of Ephrinb1 S Modulation Of Download Scientific Diagram

Utilization Of The Soft Agar Colony Formation Assay To Identify Inhibitors Of Tumorigenicity In Breast Cancer Cells Protocol

Colony Formation And Cell Growth In Cwr22rv1 Cells Treated With Egcg Download Scientific Diagram

A Mts Assay B Quantification Of Colony Formation Assay P Download Scientific Diagram
Q Tbn And9gctmvp9fr7qyyqokcnx9gi48bab1whgbinevei1czx Lrcii0wqm Usqp Cau
Plos One Shrna Targeted Commd7 Suppresses Hepatocellular Carcinoma Growth

A Quantification Of Colonies Formed In Low Density Seeding Assay Download Scientific Diagram
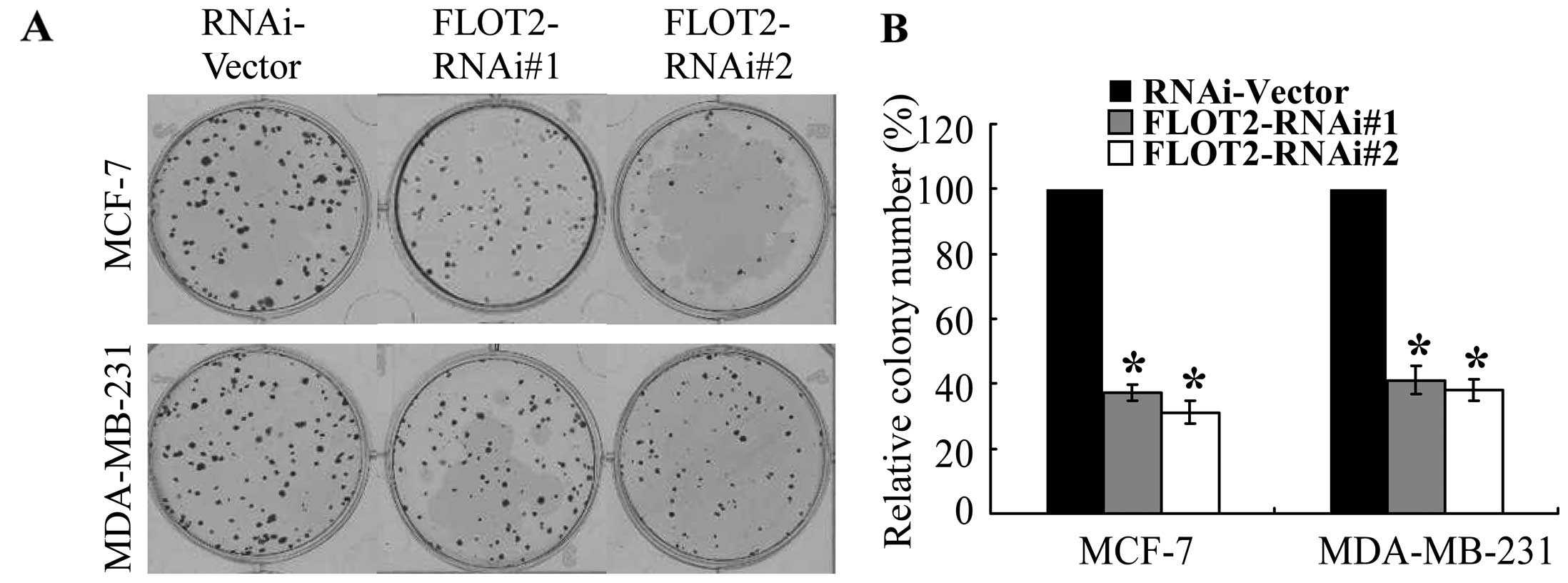
Knockdown Of Flotillin 2 Impairs The Proliferation Of Breast Cancer Cells Through Modulation Of Akt Foxo Signaling
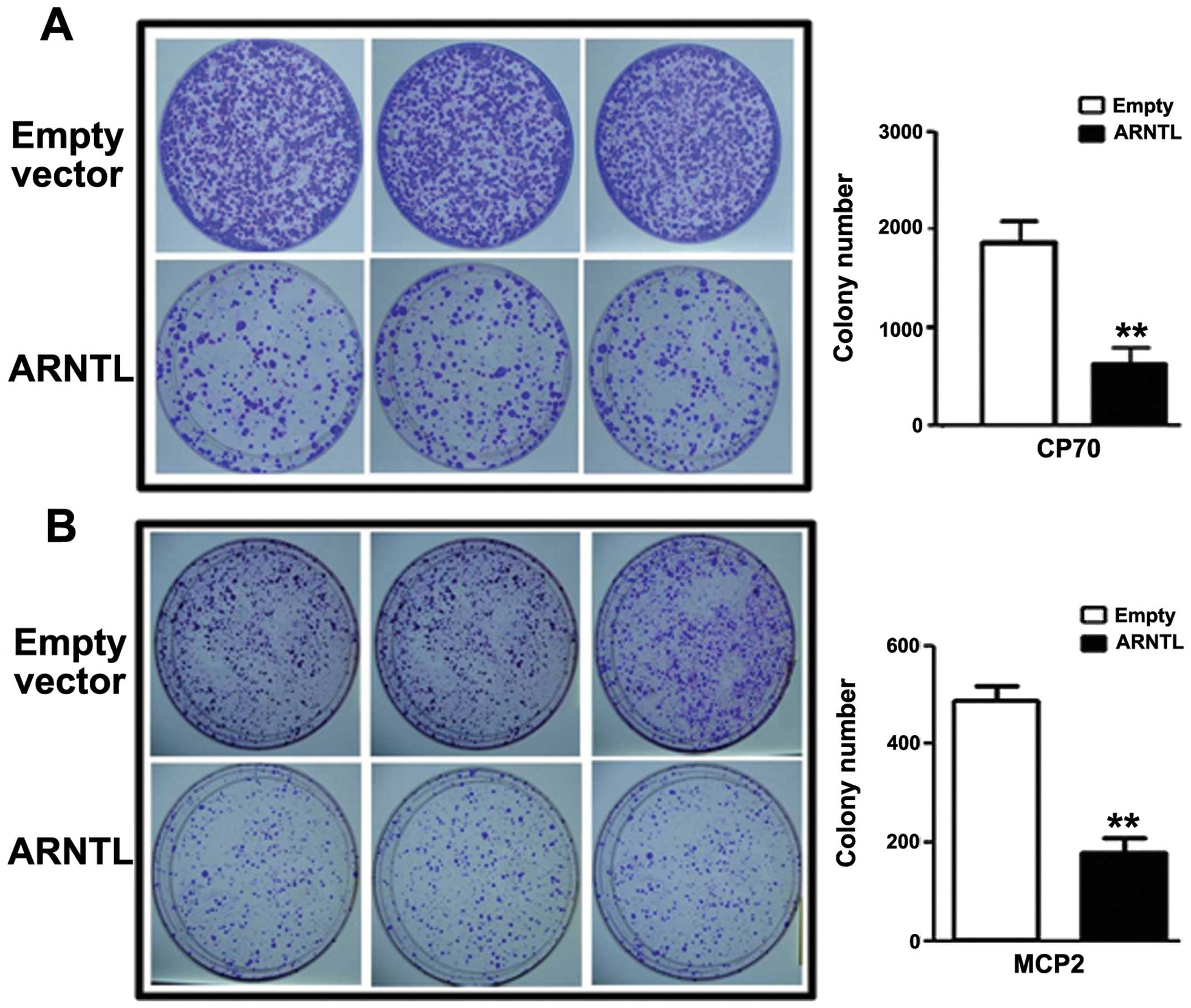
Epigenetic Silencing Of Arntl A Circadian Gene And Potential Tumor Suppressor In Ovarian Cancer
Cytosmart Clonogenic Colony Formation Assay For Stem Cells
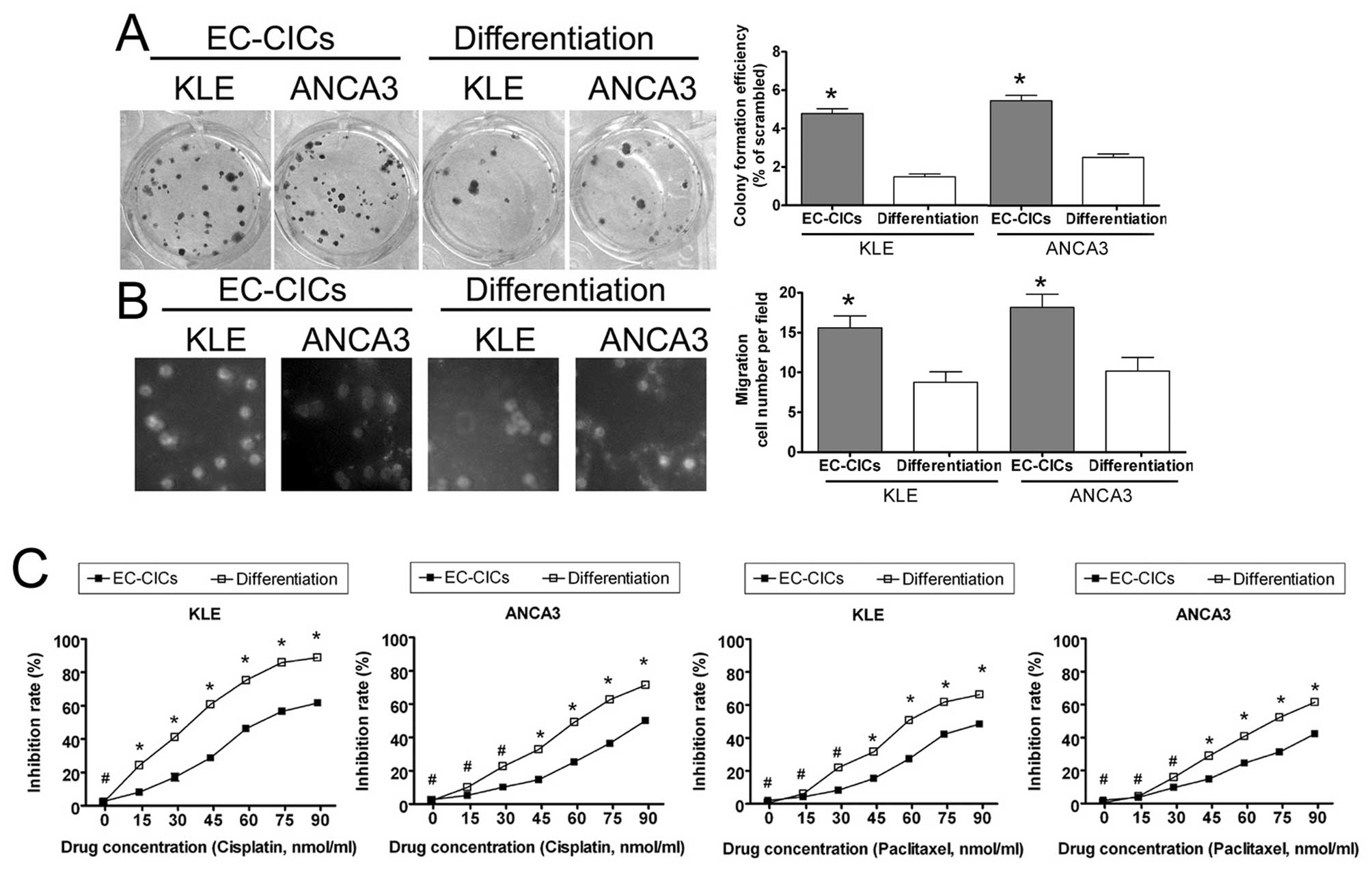
Isolation And Characterization Of Proliferative Migratory And Multidrug Resistant Endometrial Carcinoma Initiating Cells From Human Type Ii Endometrial Carcinoma Cell Lines

Figure 2 From Small Interfering Rna Sirna Mediated Knockdown Of Notch1 Suppresses Tumor Growth And Enhances The Effect Of Il 2 Immunotherapy In Malignant Melanoma Semantic Scholar
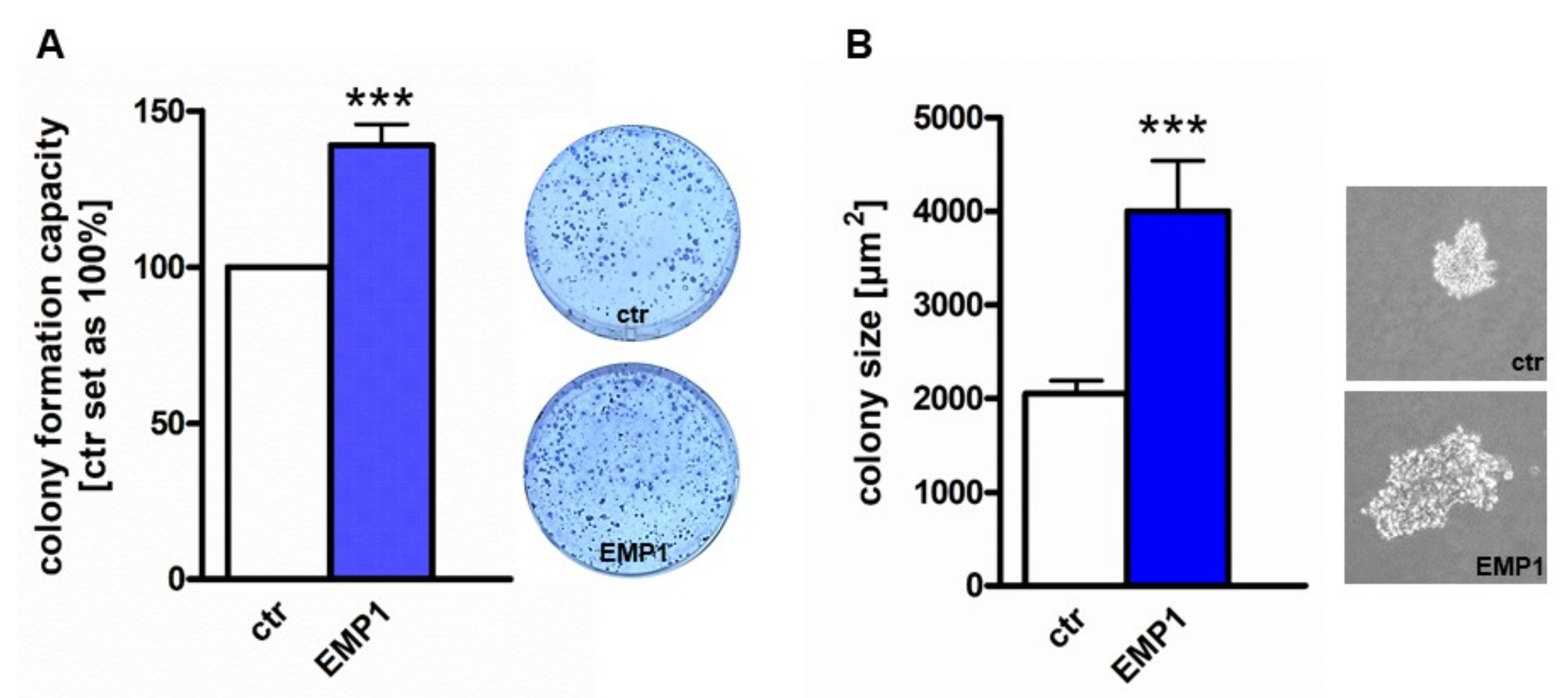
Ijms Free Full Text P53 Mir 34a And Emp1 Newly Identified Targets Of Tff3 Signaling In Y79 Retinoblastoma Cells Html

Cytosmart Clonogenic Assay What Why And How

Full Text Paeoniflorin Suppresses Pancreatic Cancer Cell Growth By Upregulating Dddt

High Throughput Fluorescent Colony Formation Assay January 10

Megacult Colony Assays Of Megakaryocyte Progenitors Cfu Mk
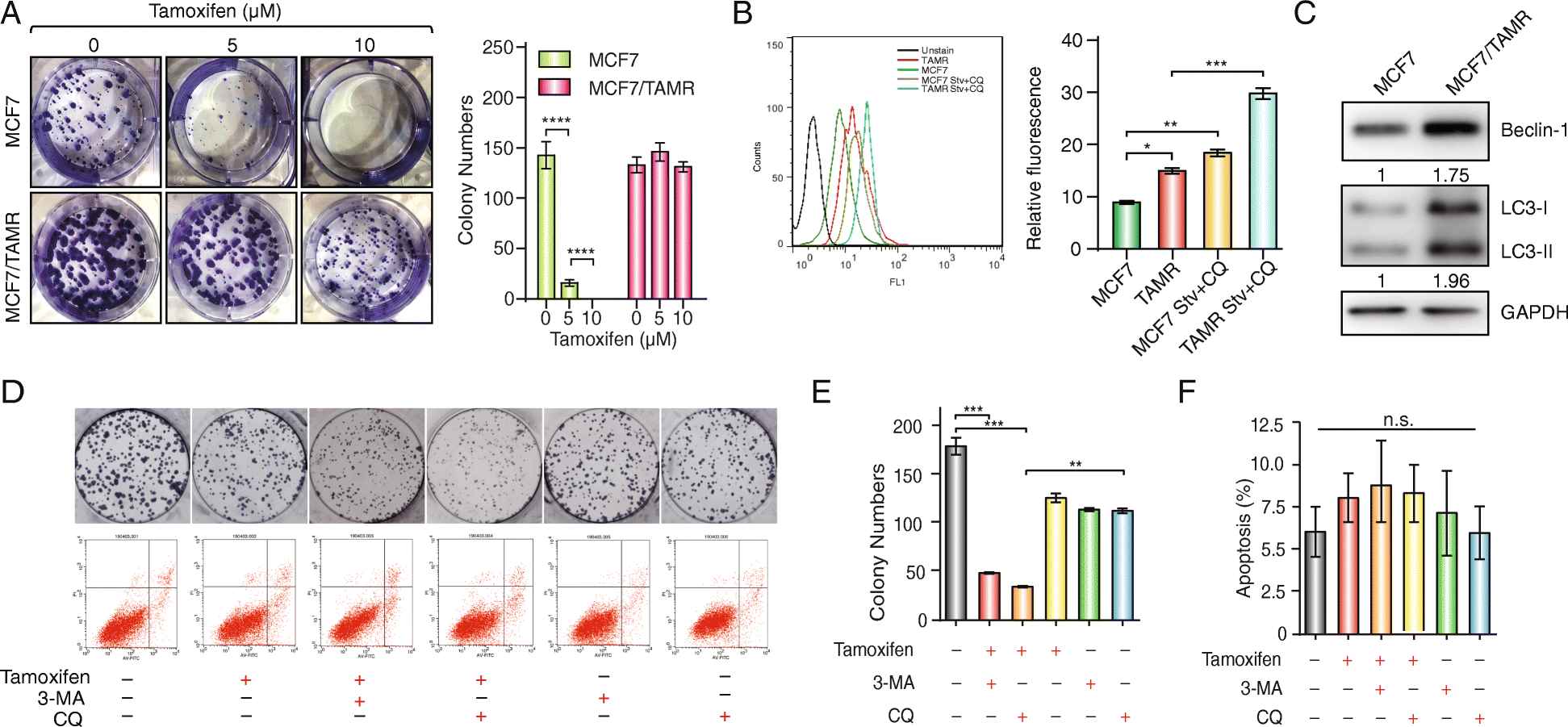
The Long Noncoding Rna H19 Promotes Tamoxifen Resistance In Breast Cancer Via Autophagy Journal Of Hematology Oncology Full Text

Cytosmart Clonogenic Assay What Why And How
Plos One Human Rad6 Promotes G1 S Transition And Cell Proliferation Through Upregulation Of Cyclin D1 Expression
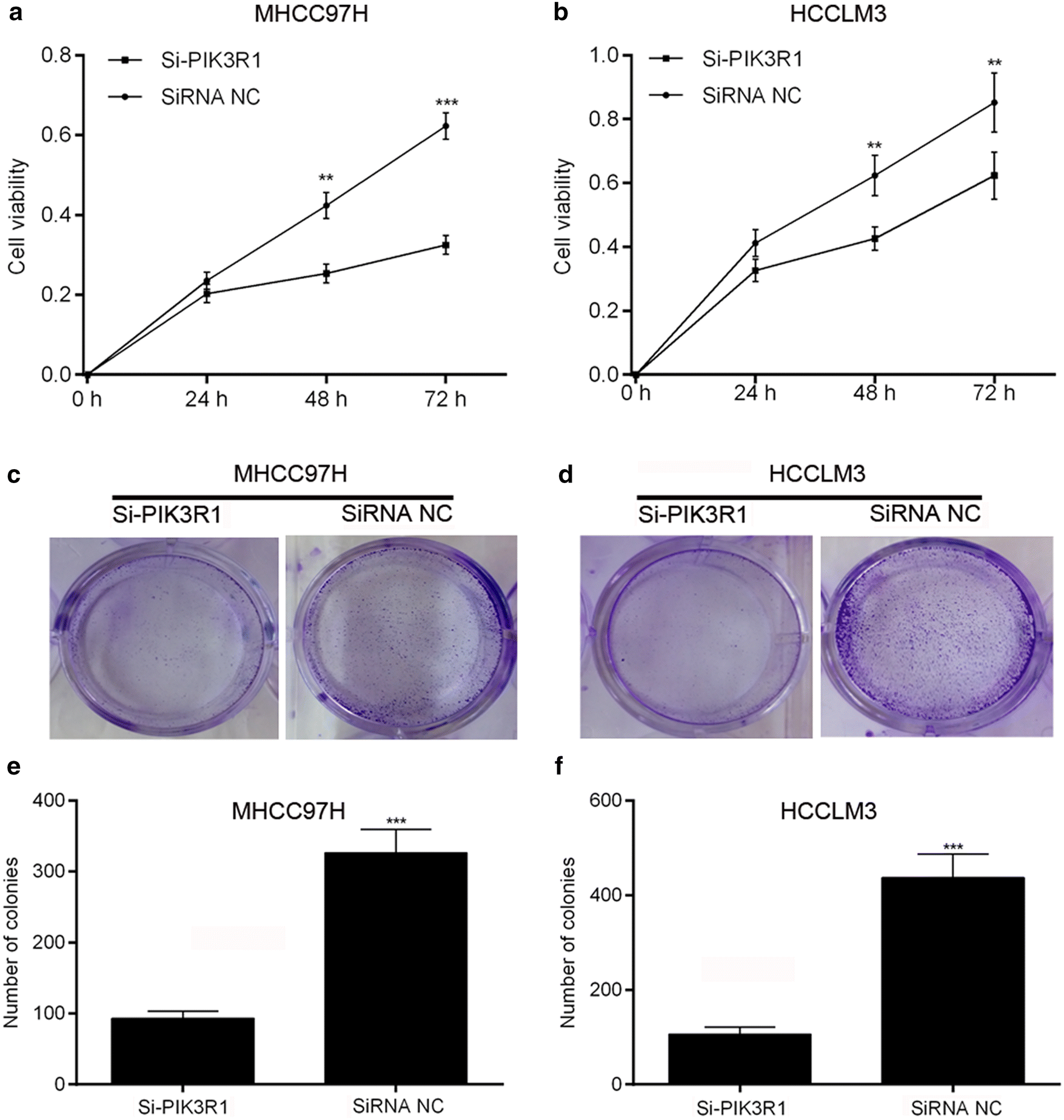
Overexpression Of Pik3r1 Promotes Hepatocellular Carcinoma Progression Biological Research Full Text

Clonogenic Assay Idea Bio Medical B

Idh1 R132h Mutation Increases Chemosensitivity To Tmz A Cell Colony Download Scientific Diagram
Q Tbn And9gcqj1jlgoj6etreht3kzoixepd Wx5pos9hqcwzu1ck Usqp Cau
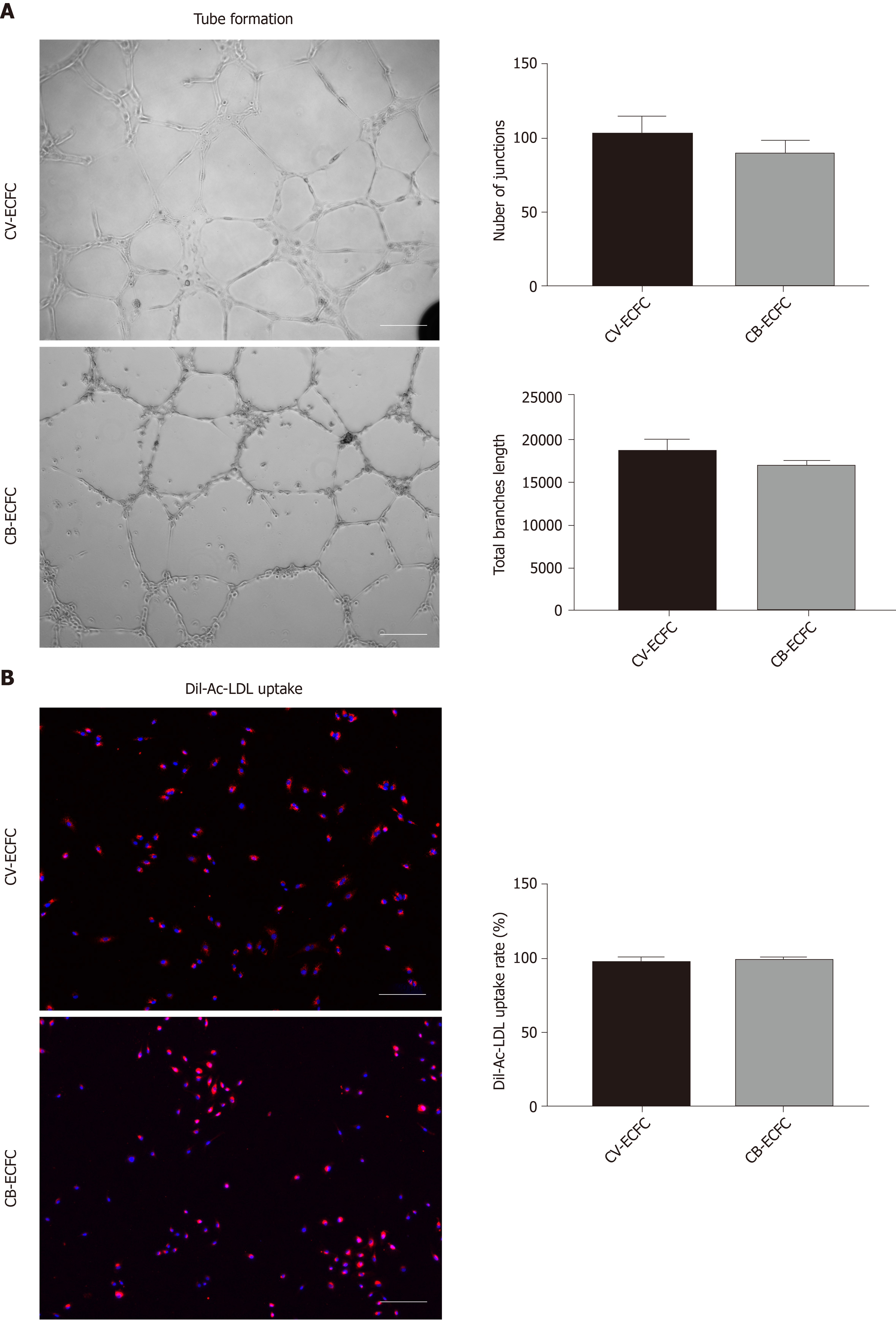
Clonal Isolation Of Endothelial Colony Forming Cells From Early Gestation Chorionic Villi Of Human Placenta For Fetal Tissue Regeneration
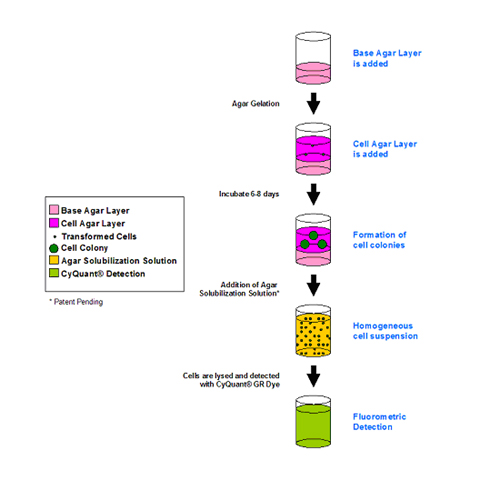
96 Well Cell Transformation Assays Standard Soft Agar
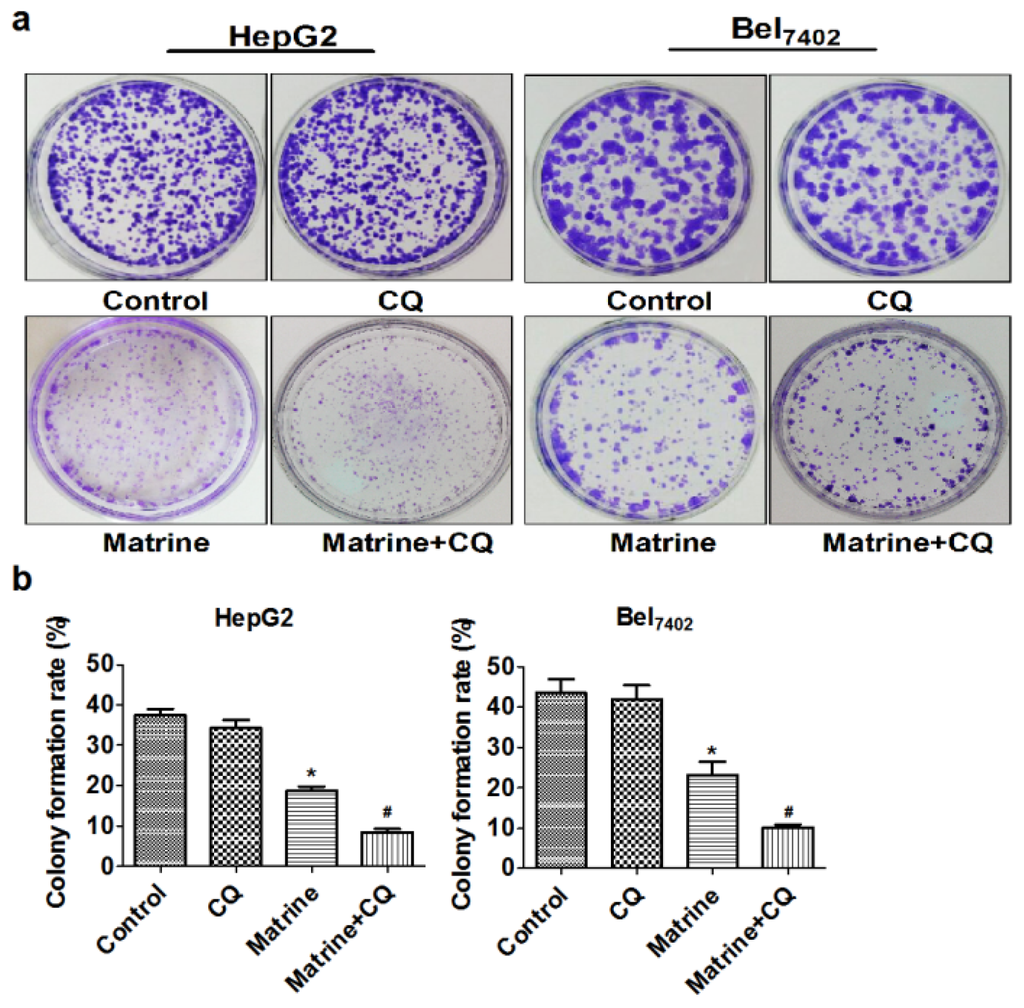
Ijms Free Full Text Blocking Autophagic Flux Enhances Matrine Induced Apoptosis In Human Hepatoma Cells Html

Oxford Optronix Gelcount

A Representative Colony Formation Assay B Quantitative Analysis Of Download Scientific Diagram

A Colony Formation Assay Representative Micrographs Left Panel And Download Scientific Diagram
Plos One Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays
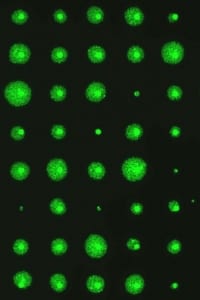
Engelward Laboratory Microcolonychip Cell Survival Quantitation

Soft Agar Colony Formation Assay Creative Bioarray Creative Bioarray

The Soft Agar Colony Formation Assay Protocol
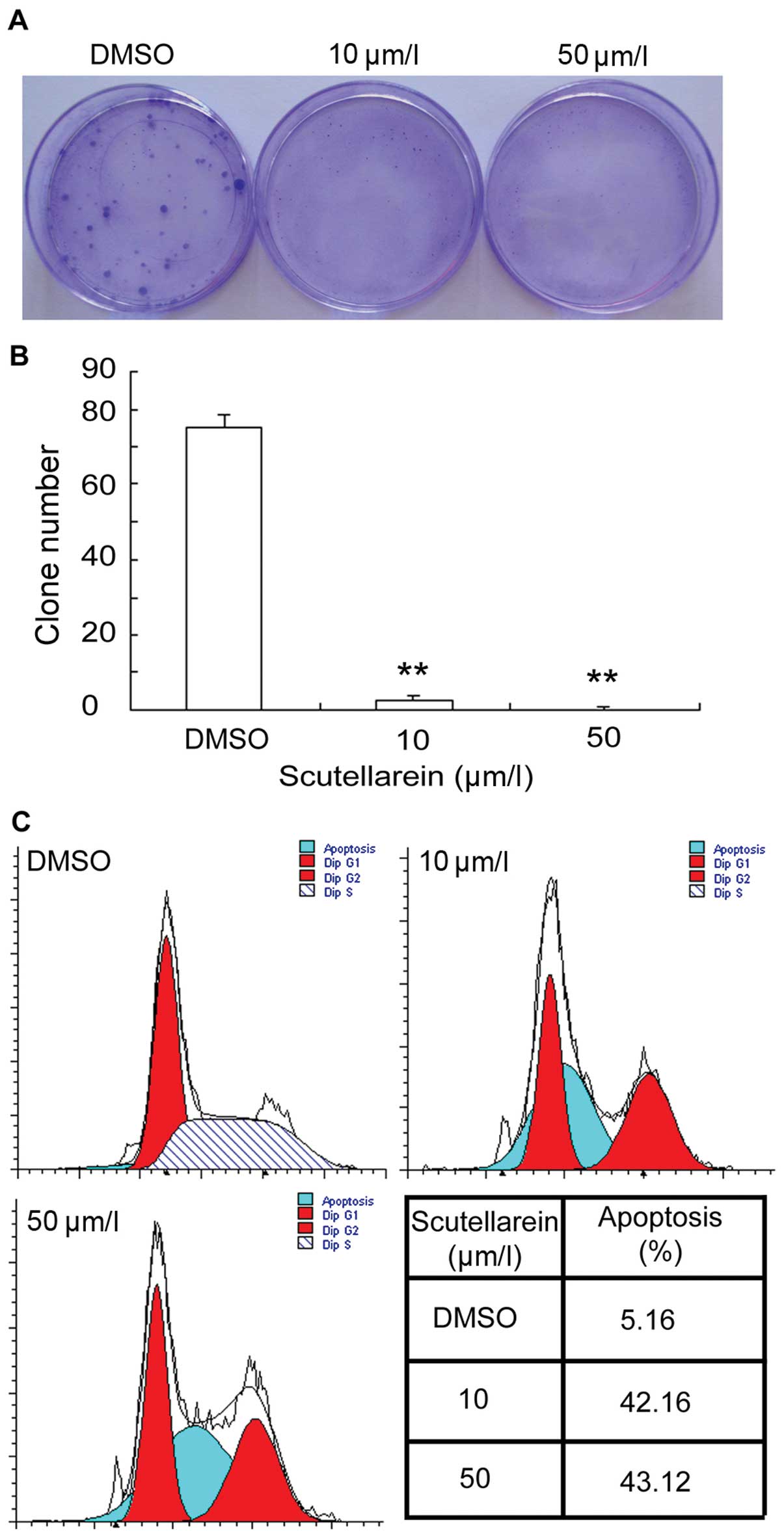
Scutellarein Inhibits Cancer Cell Metastasis In Vitro And Attenuates The Development Of Fibrosarcoma In Vivo
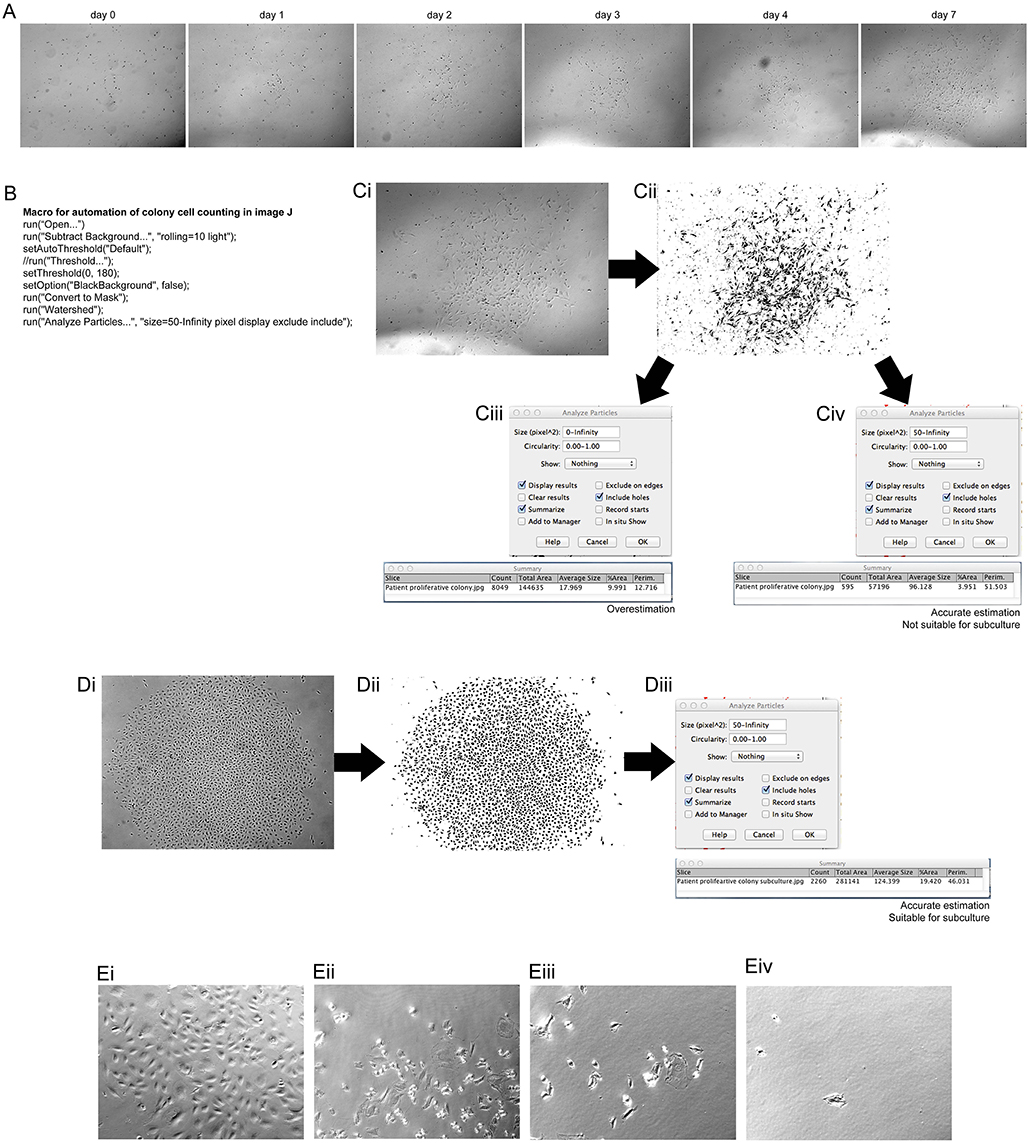
Frontiers The Use Of Live Cell Imaging And Automated Image Analysis To Assist With Determining Optimal Parameters For Angiogenic Assay In Vitro Cell And Developmental Biology

Pdf Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays Semantic Scholar

Clonogenic Assay An Overview Sciencedirect Topics

A B Representative Images And Quantification Of Colony Forming Assay Download Scientific Diagram

Cytosmart Clonogenic Colony Formation Assay For Stem Cells

An Improved Crystal Violet Assay For Biofilm Quantification In 96 Well Microtitre Plate Biorxiv
Www 2bscientific Com Getmedia 48cf7444 f3 4bbe B1ca Df2df3e965 Colony Formation Brochure 14 Pdf
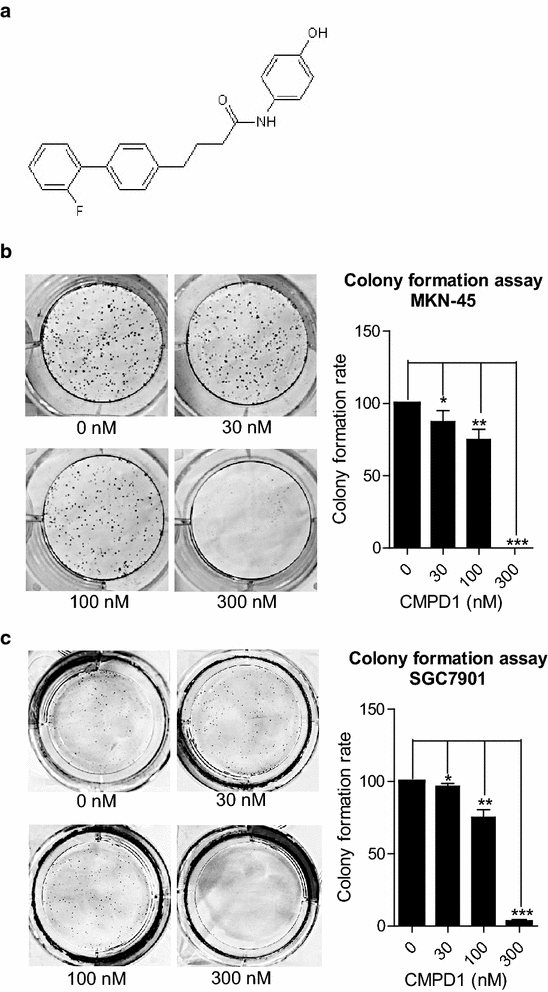
Cmpd1 Inhibited Human Gastric Cancer Cell Proliferation By Inducing Apoptosis And G2 M Cell Cycle Arrest Biological Research Full Text
Www Cell Com Cell Reports Pdf S2211 1247 19 300 8 Pdf

Cytosmart Clonogenic Assay What Why And How
Plos One The Histone Demethylase Jarid1b Kdm5b Is A Novel Component Of The Rb Pathway And Associates With E2f Target Genes In Mefs During Senescence
Www Cell Com Cell Reports Pdfextended S2211 1247 19 300 8
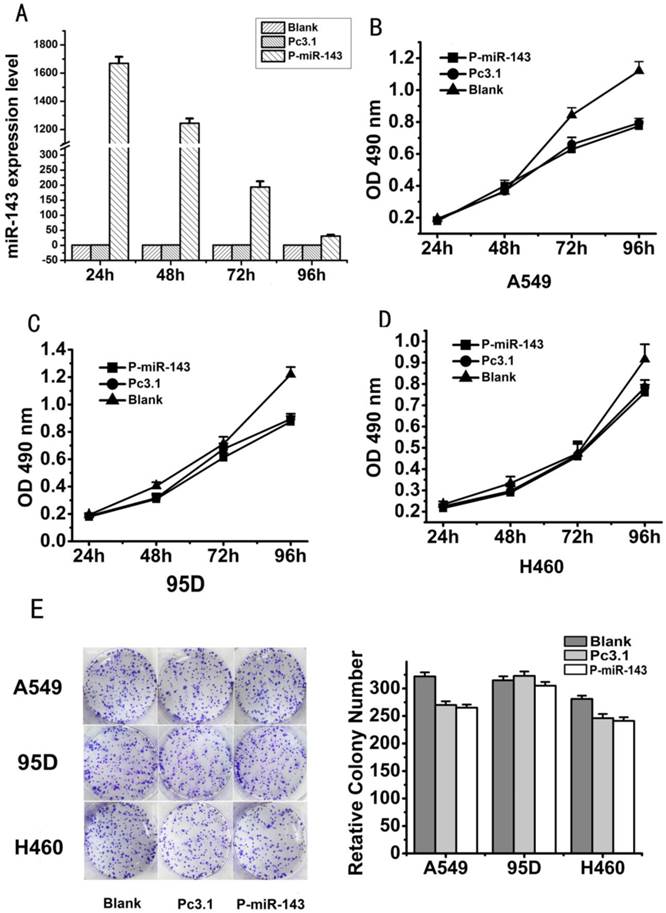
Microrna 143 Inhibits Migration And Invasion Of Human Non Small Cell Lung Cancer And Its Relative Mechanism
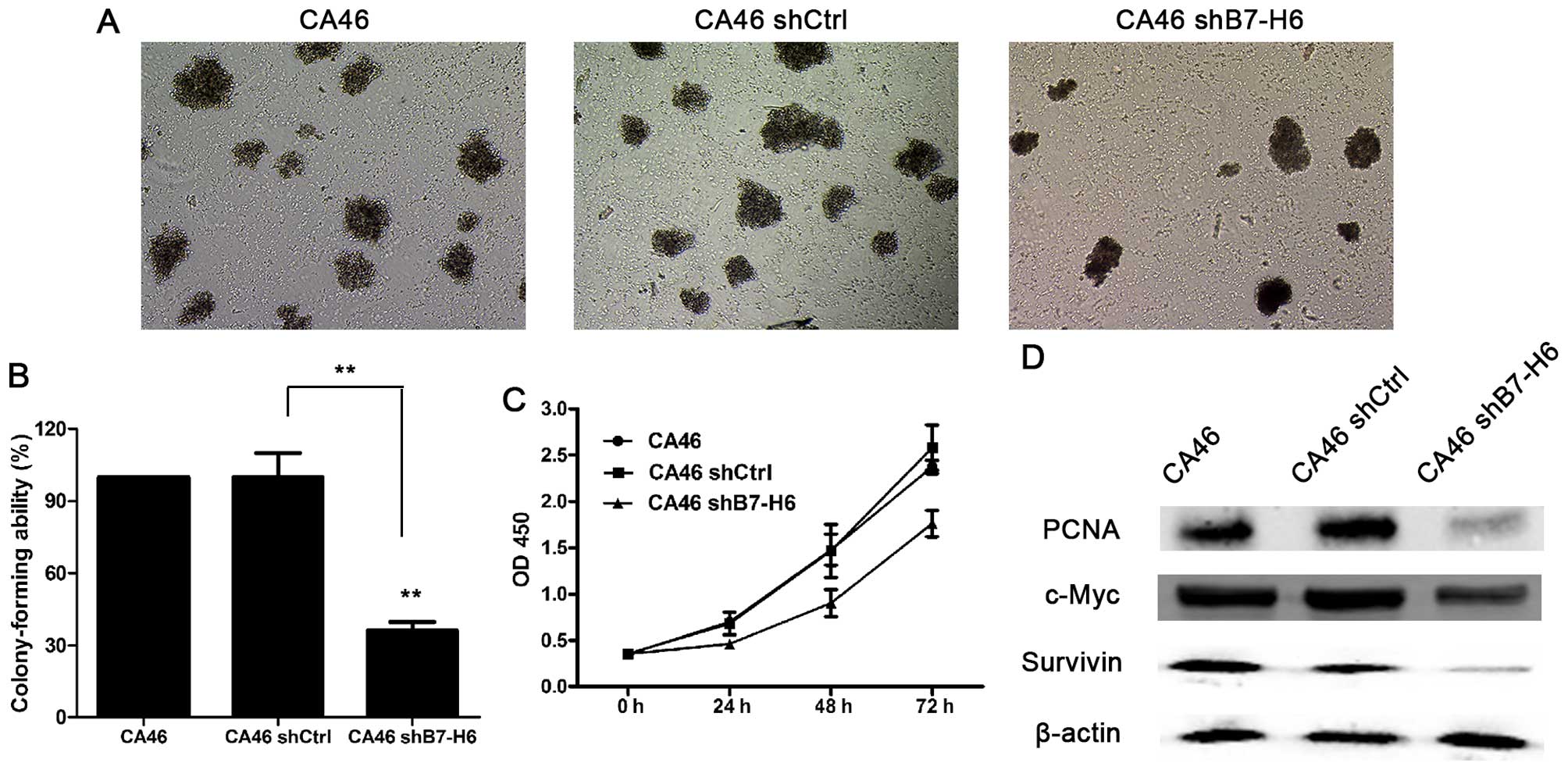
Knockdown Of H6 Inhibits Tumor Progression And Enhances Chemosensitivity In B Cell Non Hodgkin Lymphoma

A The Effects On Colony Formation Of Hepg2 Cells Treated With Pbs Download Scientific Diagram

Clonogenic Assay Of Nanoparticles Or Free Dox Or Elc In Hepg2 Cells Download Scientific Diagram
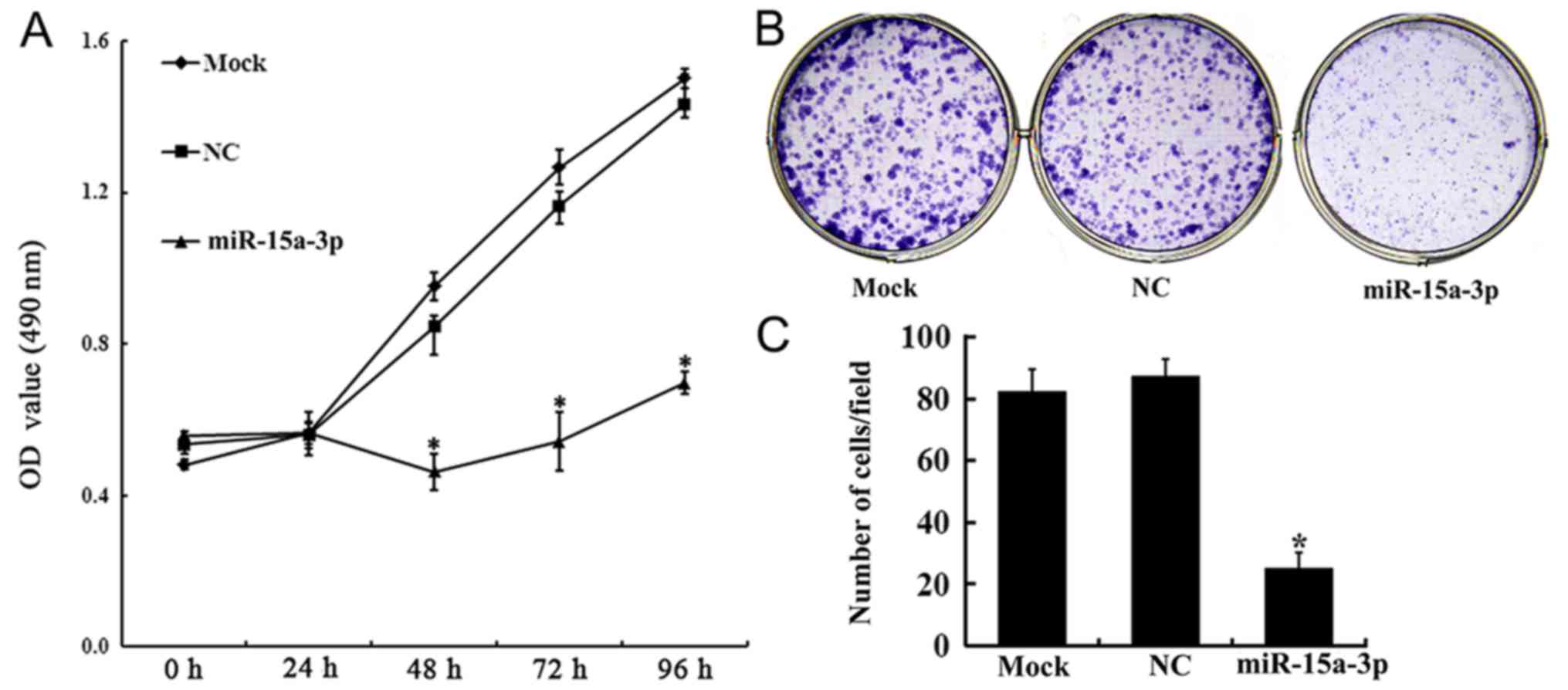
Mir 15a 3p Affects The Proliferation Migration And Apoptosis Of Lens Epithelial Cells

High Throughput Fluorescent Colony Formation Assay January 10
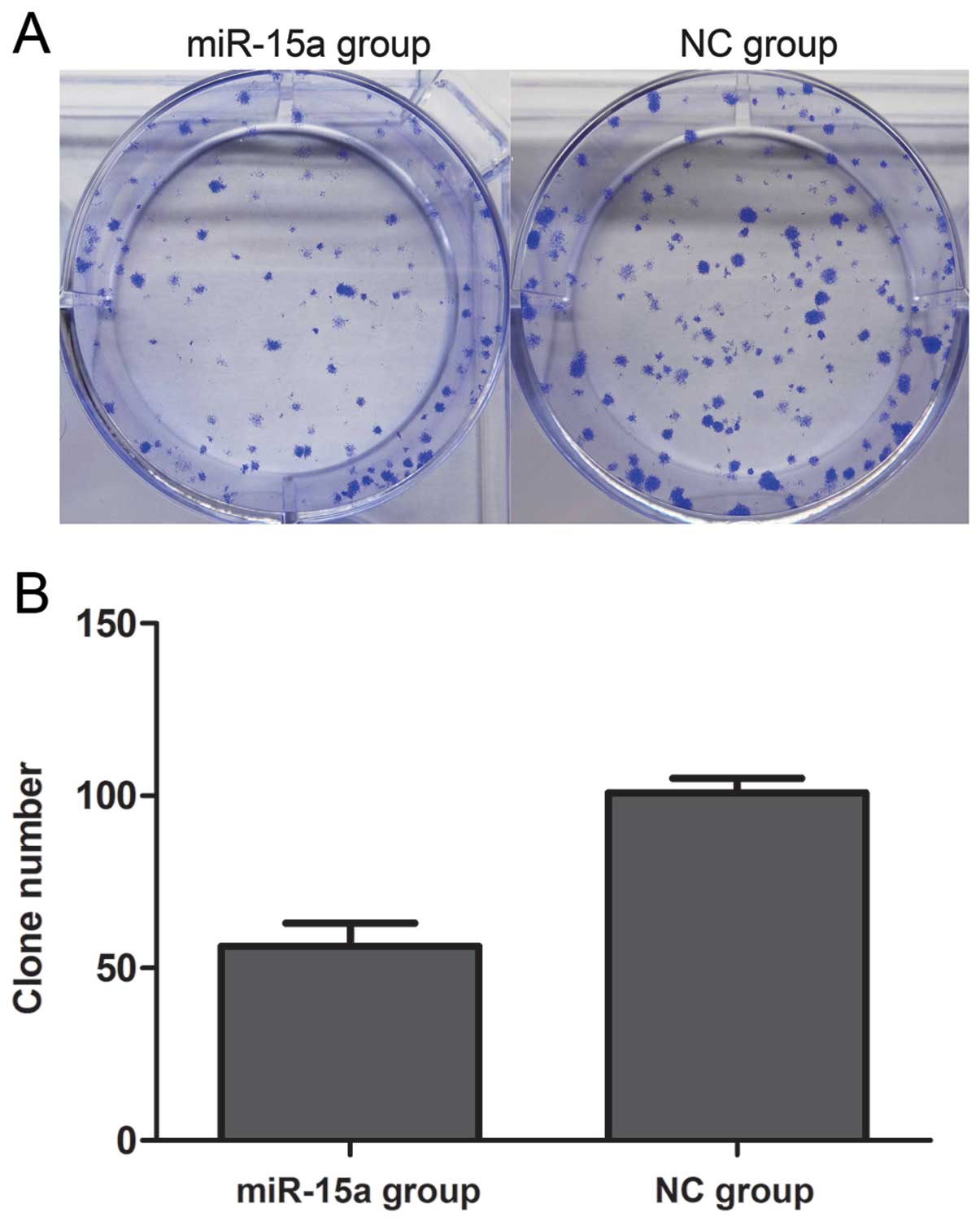
Mir 15a Is Underexpressed And Inhibits The Cell Cycle By Targeting Ccne1 In Breast Cancer
Plos One Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays

Clonogenic Assay Youtube
Plos One Colonyarea An Imagej Plugin To Automatically Quantify Colony Formation In Clonogenic Assays
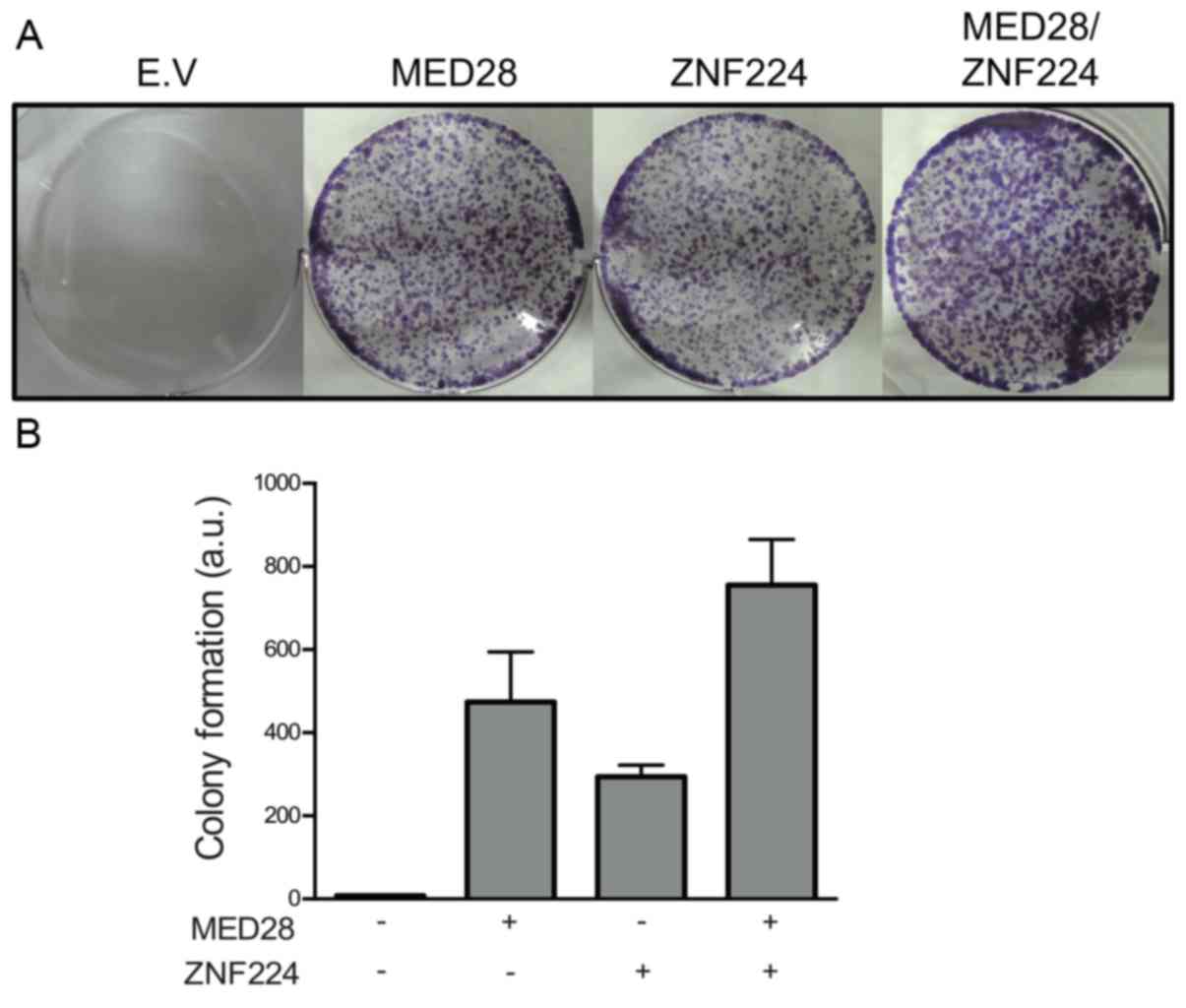
Med28 Increases The Colony Forming Ability Of Breast Cancer Cells By Stabilizing The Znf224 Protein Upon Dna Damage

High Throughput Fluorescent Colony Formation Assay Metrolab Blog

High Throughput Fluorescent Colony Formation Assay January 10
Www 2bscientific Com Getmedia 48cf7444 f3 4bbe B1ca Df2df3e965 Colony Formation Brochure 14 Pdf

Metformin Inhibits Tumorigenesis And Tumor Growth Of Breast Cancer Cells By Upregulating Mir 0c But Downregulating Akt2 Expression
Plos One The Architectural Chromatin Factor High Mobility Group A1 Enhances Dna Ligase Iv Activity Influencing Dna Repair

A The Fbxw7 Mrna Expression Level Of Hcc Cell Lines N 3 B Colony Download Scientific Diagram

Culture And Analysis Of Hematopoietic Progenitor Cells In Cfu Assays Stemcell Technologies

Clonogenic Assay Creative Bioarray Creative Bioarray
Cytosmart Clonogenic Colony Formation Assay For Stem Cells
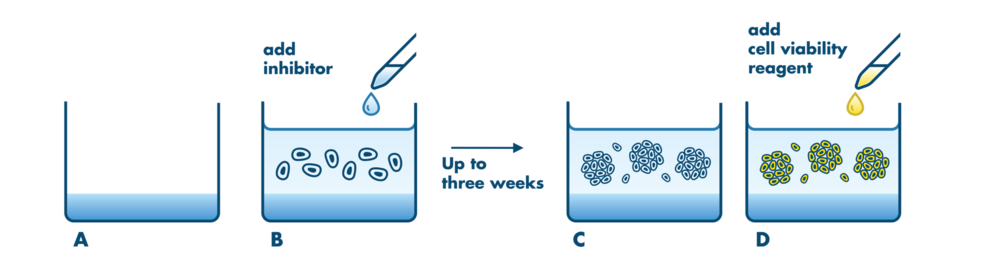
Cell Based Soft Agar Assays Reaction Biology
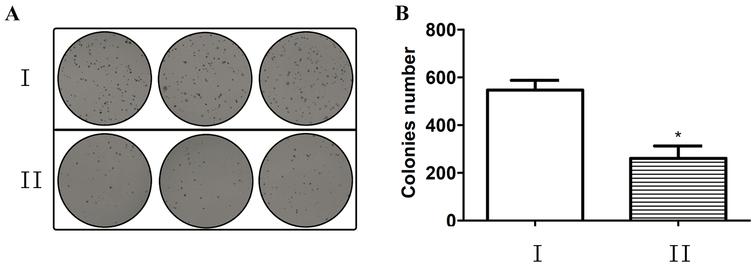
Inhibition Of Mir 155 A Therapeutic Target For Breast Cancer Prevented In Cancer Stem Cell Formation Ios Press

Brd4 Is Dispensable In Naive Escs A Quantification Of The Colony Download Scientific Diagram
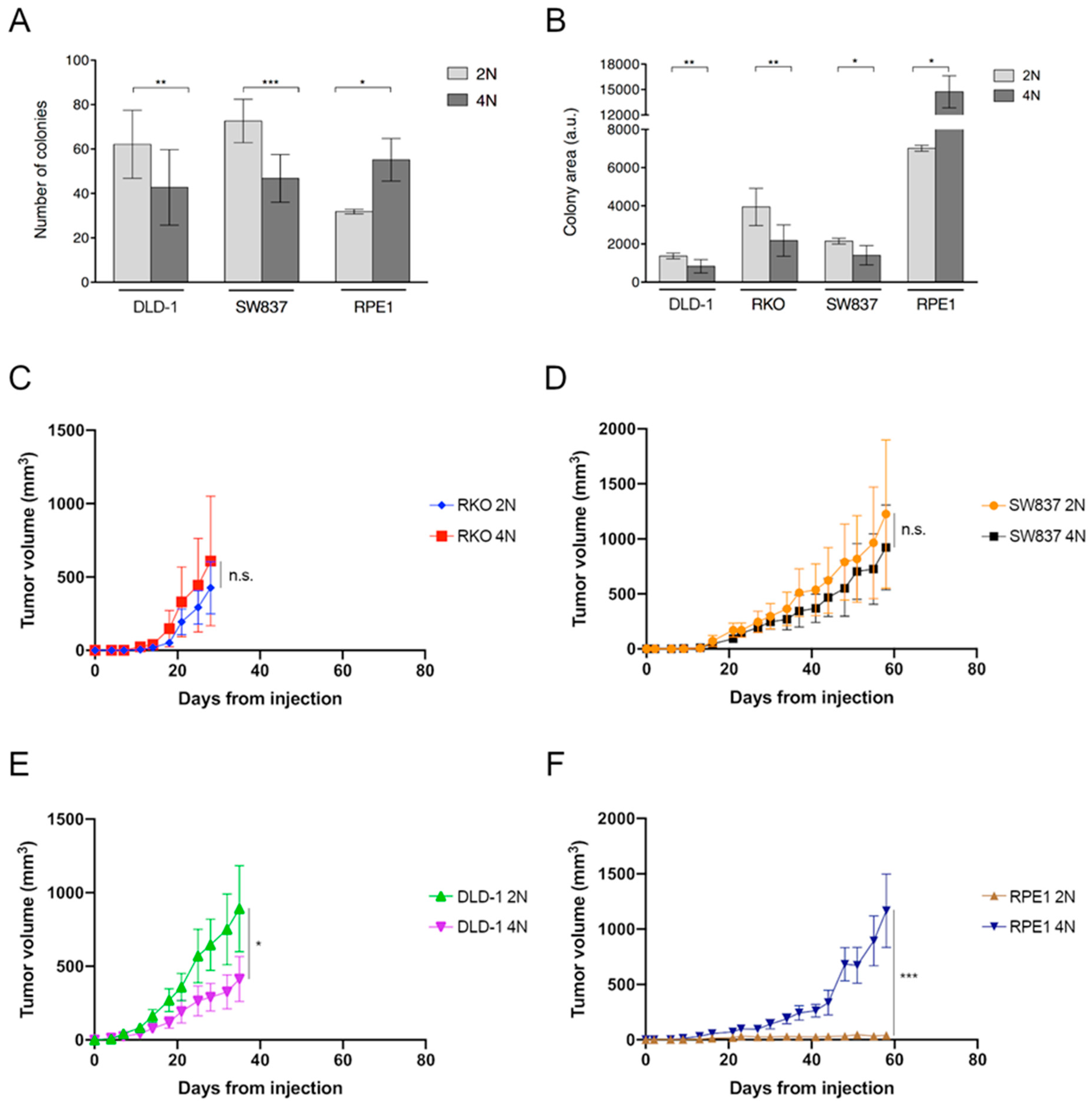
Cancers Free Full Text Tetraploidy Associated Genetic Heterogeneity Confers Chemo Radiotherapy Resistance To Colorectal Cancer Cells Html
Www 2bscientific Com Getmedia 48cf7444 f3 4bbe B1ca Df2df3e965 Colony Formation Brochure 14 Pdf

Figure 2 S I Ammson I Is Required For Melanoma Growth And Survival

Comparison Of The Colony Formation And Crystal Violet Cell Proliferation Assays To Determine Cellular Radiosensitivity In A Repair Deficient Mcf10a Cell Line Sciencedirect
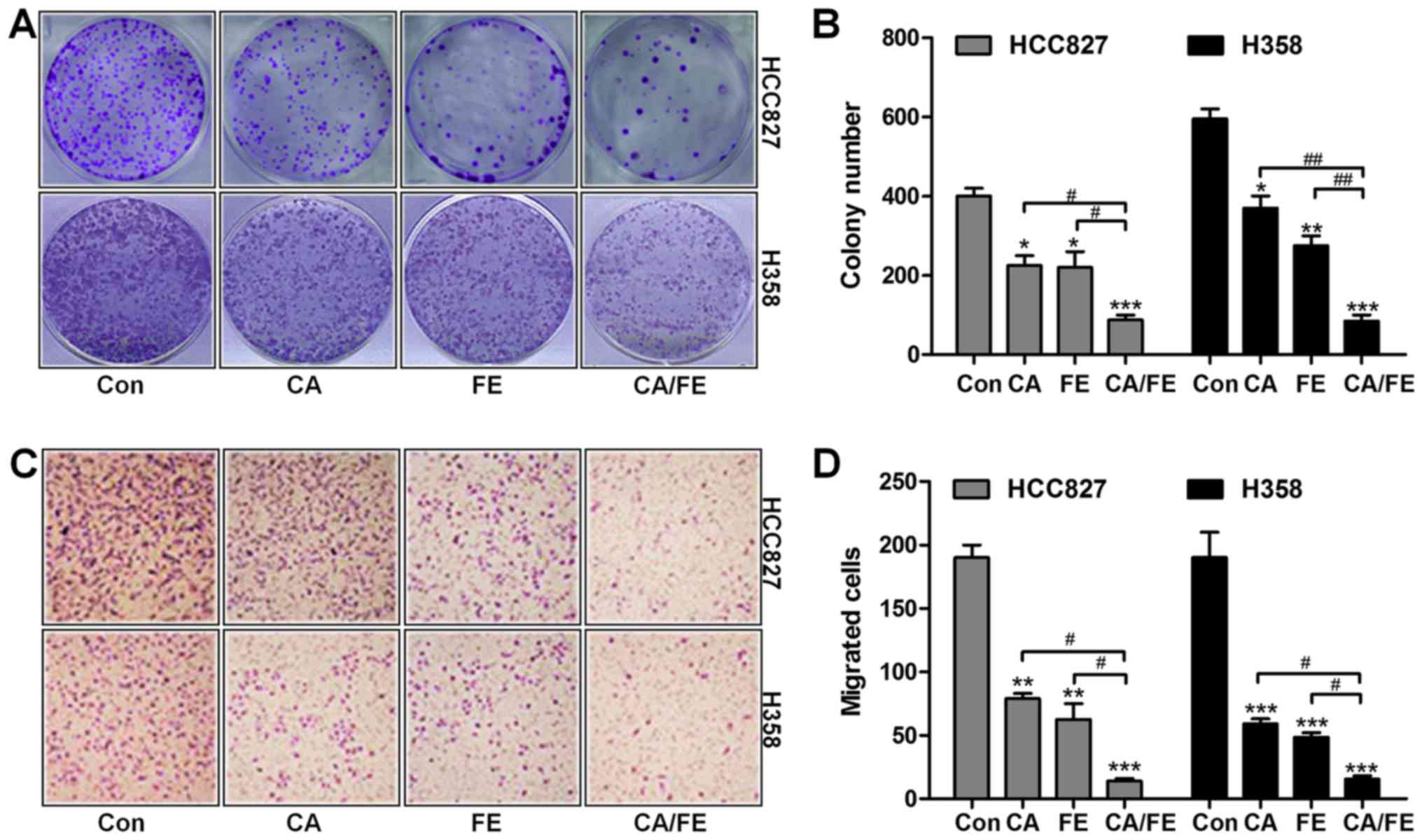
Carnosic Acid And Fisetin Combination Therapy Enhances Inhibition Of Lung Cancer Through Apoptosis Induction

Cytosmart Clonogenic Assay What Why And How

Cytosmart Clonogenic Assay What Why And How

Tumor Spheroid Formation Assay Sigma Aldrich
Q Tbn And9gcslemxvf V3xbbavhlwxx360golzqpnf4bfn Jgeeabr9mtoz9k Usqp Cau

Imagej Macros For The User Friendly Analysis Of Soft Agar And Wound Healing Assays Biotechniques



